

Let's explore Behind the CODE
This site features a collection of projects I've contributed to, including open-source contributions, APIs, large-scale data analysis, and scalable systems.

Crosslinking
Advanced protein crosslinking analysis tool.
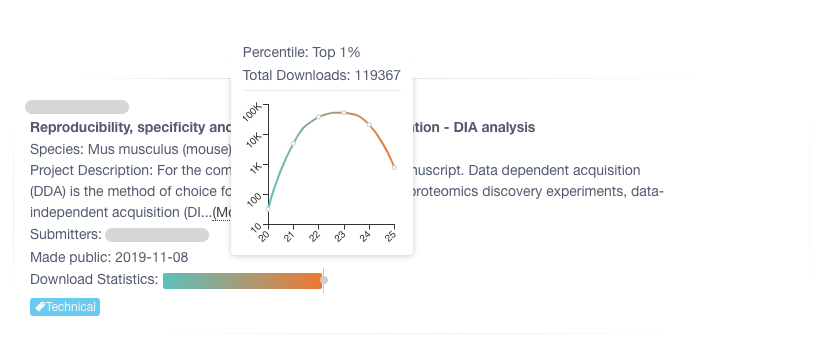
Nextflow File Download Statistics Pipeline
Pipeline to analyze file download statistics.
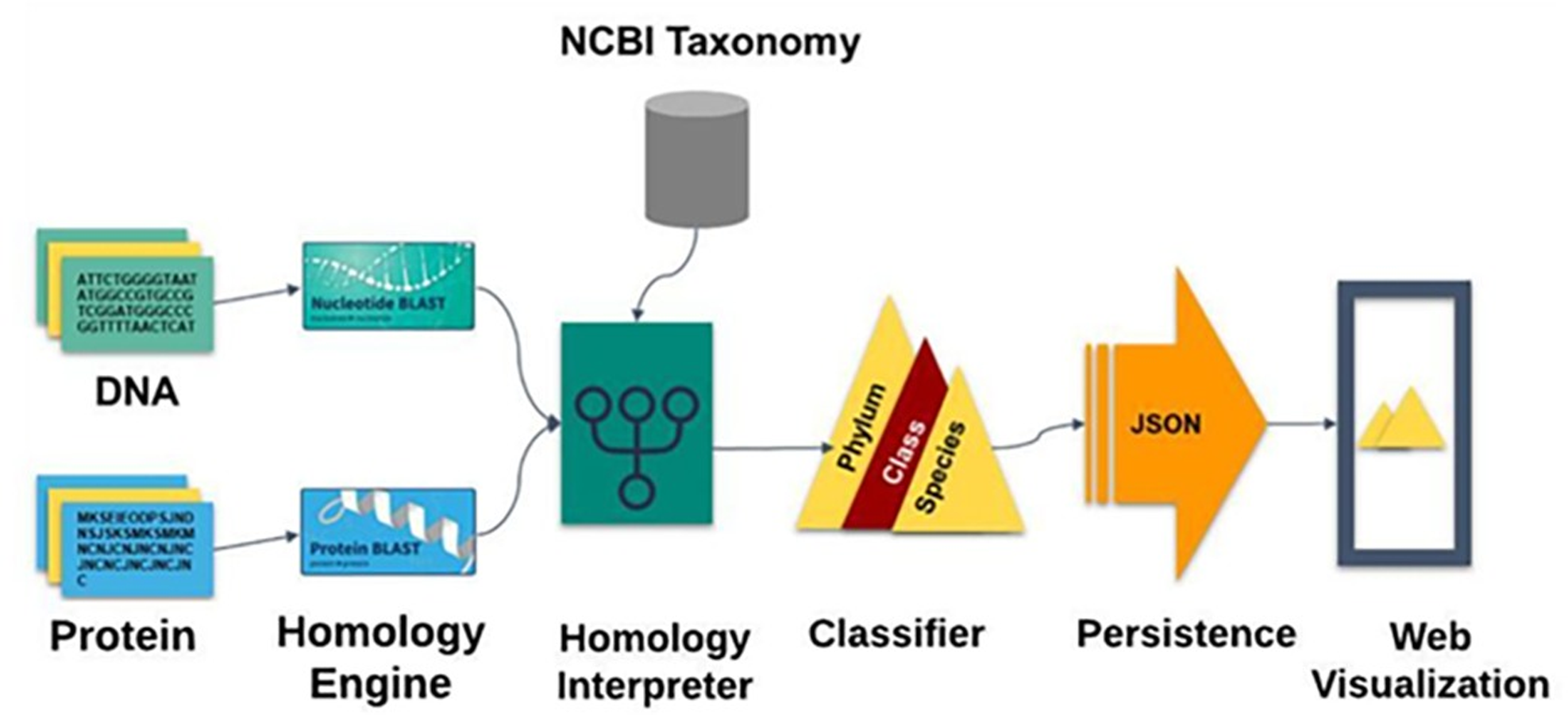
Orfan-genes
Many sequenced genomes reveal that a significant percentage of genes in a given organism's taxon lack orthologous sequences in other taxa. These are called "orphans" or "ORFans" if present in one species or "taxonomically restricted genes" (TRGs) at higher levels. Research on these genes is key to understanding their origins, but current software for identifying them is limited, complex, and has restricted database access. ORFanID is a user-friendly web-based tool that efficiently identifies orphan genes and TRGs across taxonomic levels, using sequences from large bioinformatics repositories like NCBI. It allows users to control search parameters and presents results in sortable, filterable tables with graphical displays of taxonomic trees.
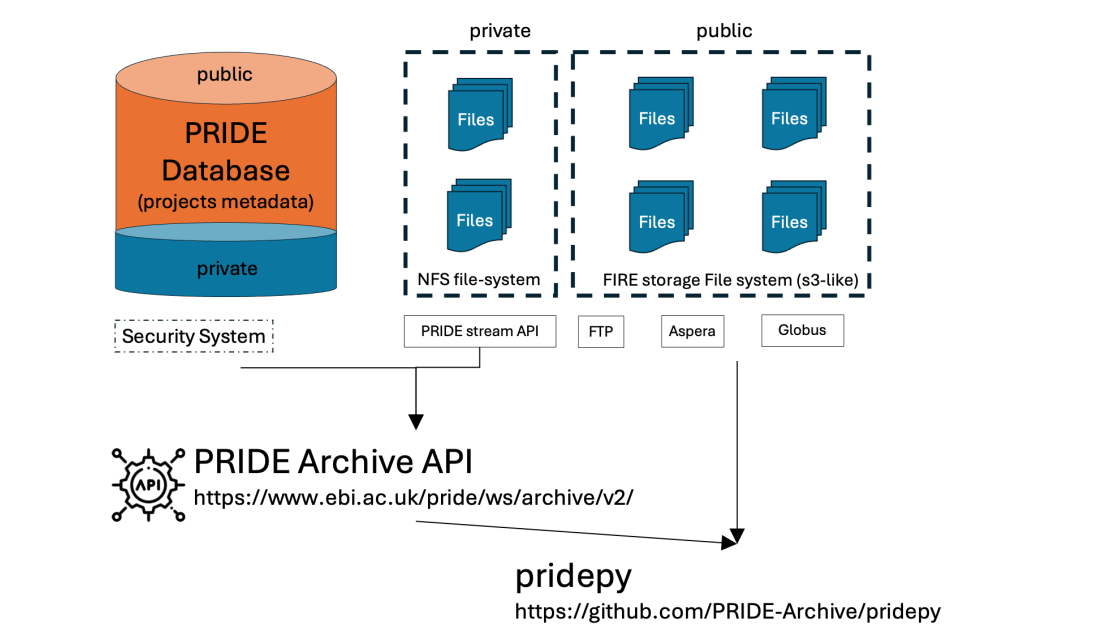
pridepy
The Proteomics Identification Database (PRIDE) is the world’s largest repository for proteomics data and a founding member of ProteomeXchange. Here, we introduce pridepy, a Python client designed to access PRIDE Archive data, including project metadata and file downloads. pridepy offers a flexible programmatic interface for searching, retrieving, and downloading data via the PRIDE REST API. This tool simplifies the integration of PRIDE datasets into bioinformatics pipelines, making it easier for researchers to handle large datasets programmatically.

GeoDiver
GeoDiver is an online web application for performing Differential Gene Expression Analysis (DGEA) and Generally Applicable Gene-set Enrichment Analysis (GAGE) on gene expression datasets from the publicly available Gene Expression Omnibus (GEO). The output produced includes numerous high quality interactive graphics, allowing users to easily explore and examine complex datasets instantly. Furthermore, the results produced can be reviewed at a later date and shared with collaborators.
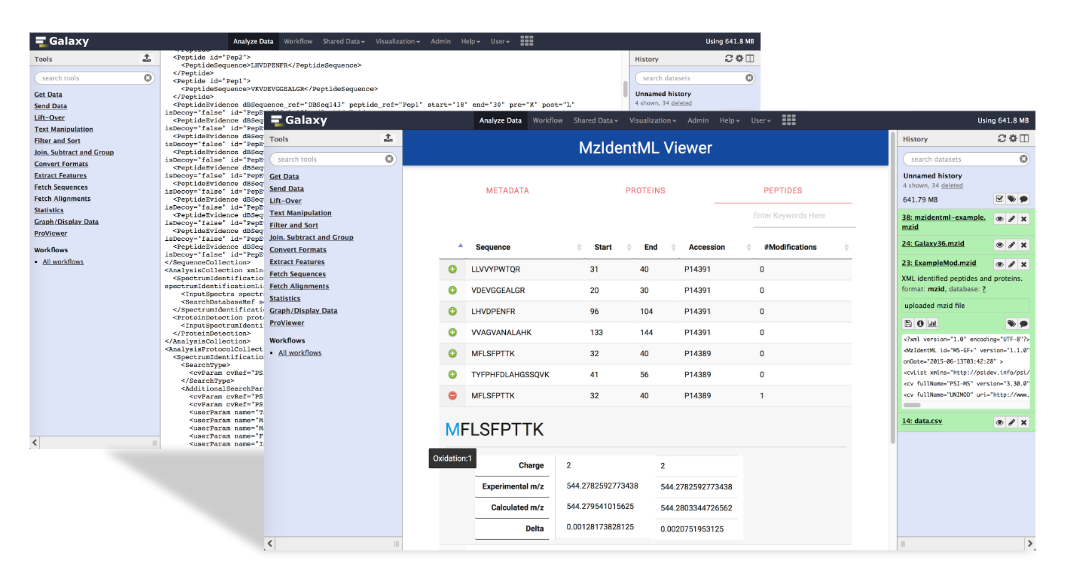
Galaxy Plugin
This project developed ProExtractor, a Java library that extracts proteomics data from mzIdentML files and converts it into a lightweight JSON format. The ProViewer plugin, built with web technologies, visualizes this data interactively in Galaxy. ProExtractor is 83% faster than MzidToHTML, and ProViewer reduces visualization delays by 98% through caching. ProViewer is integrated into Galaxy and available on the public GIO Galaxy server, with an easy installation script for custom servers.
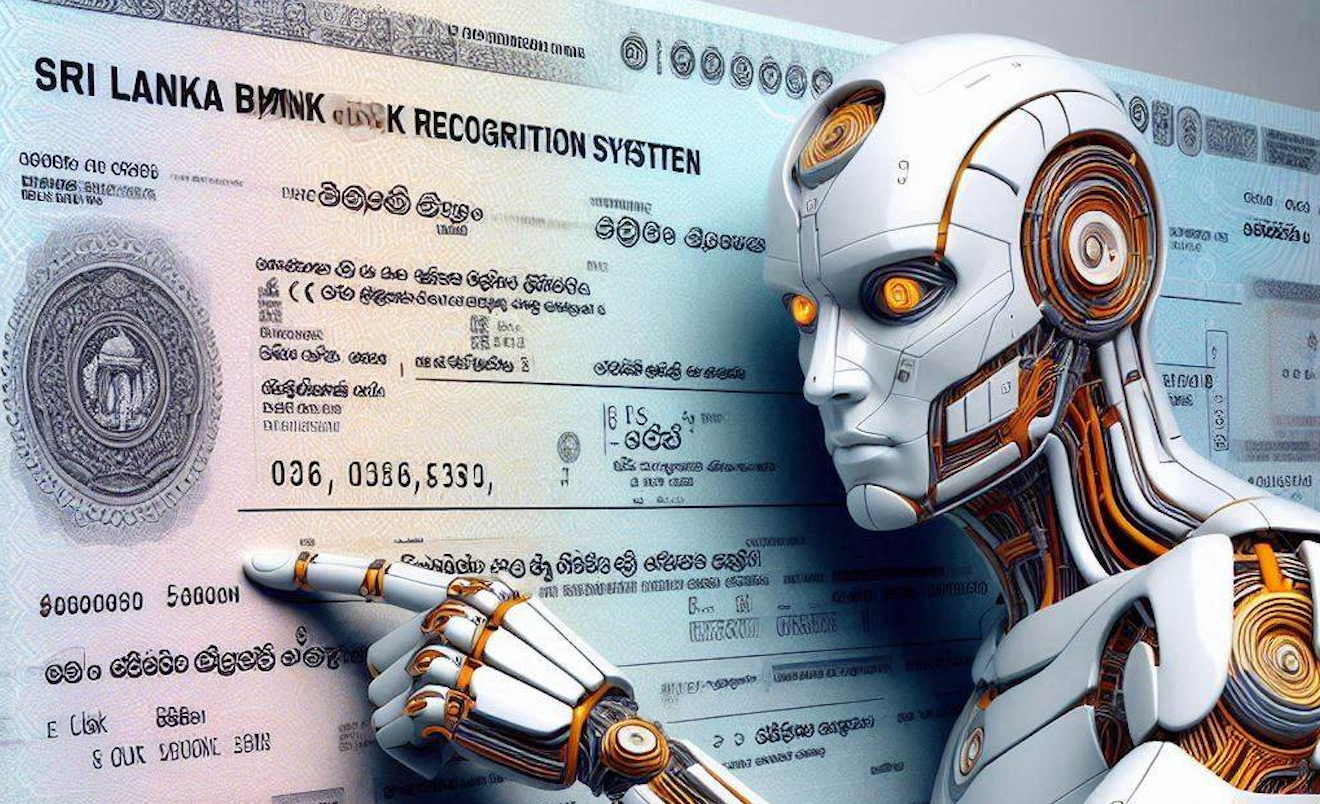
Automated Bank Cheque Recongition System using OCR
Automated bank cheque details extraction using Optical Charactor Recongition system. It was recongising Bank No, Account No which are printed charactors 99%, handwritten date recognised 80% and handwritten amount in letters and numbers 63%.
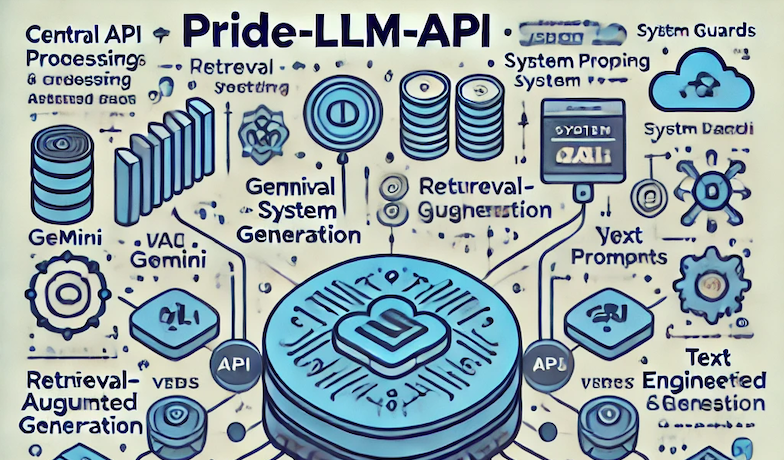
PRIDE-AI-CORE
The Pride-LLM-API is a modular, scalable, and secure API designed for integrating LLM-powered functionalities with structured data storage and retrieval. Built in Python, the project follows a clean architecture with well-defined components.This project act as the base API for AI-driven document processing, retrieval-augmented generation (RAG), and secure NLP applications.
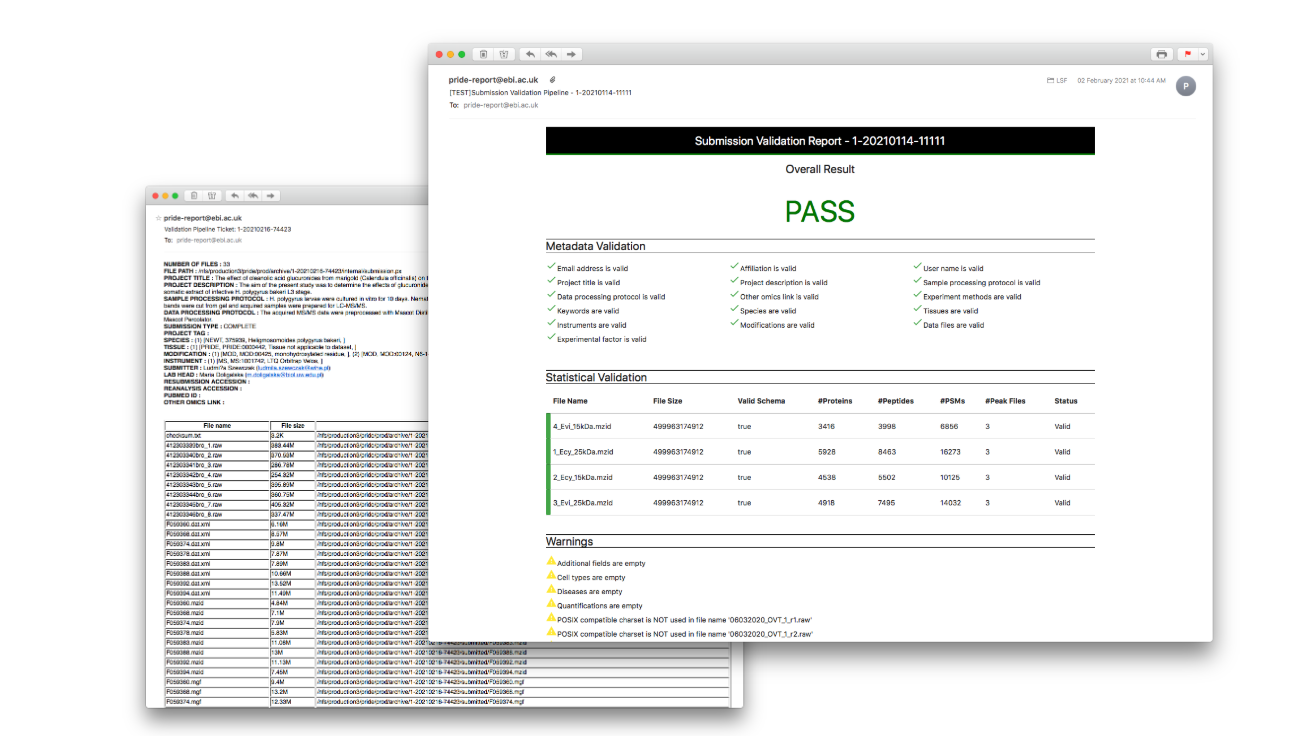
Nextflow Validation Pipeline
An efficient nextflow validation pipeline was developed to solve some of the issues in current production validation pipeline
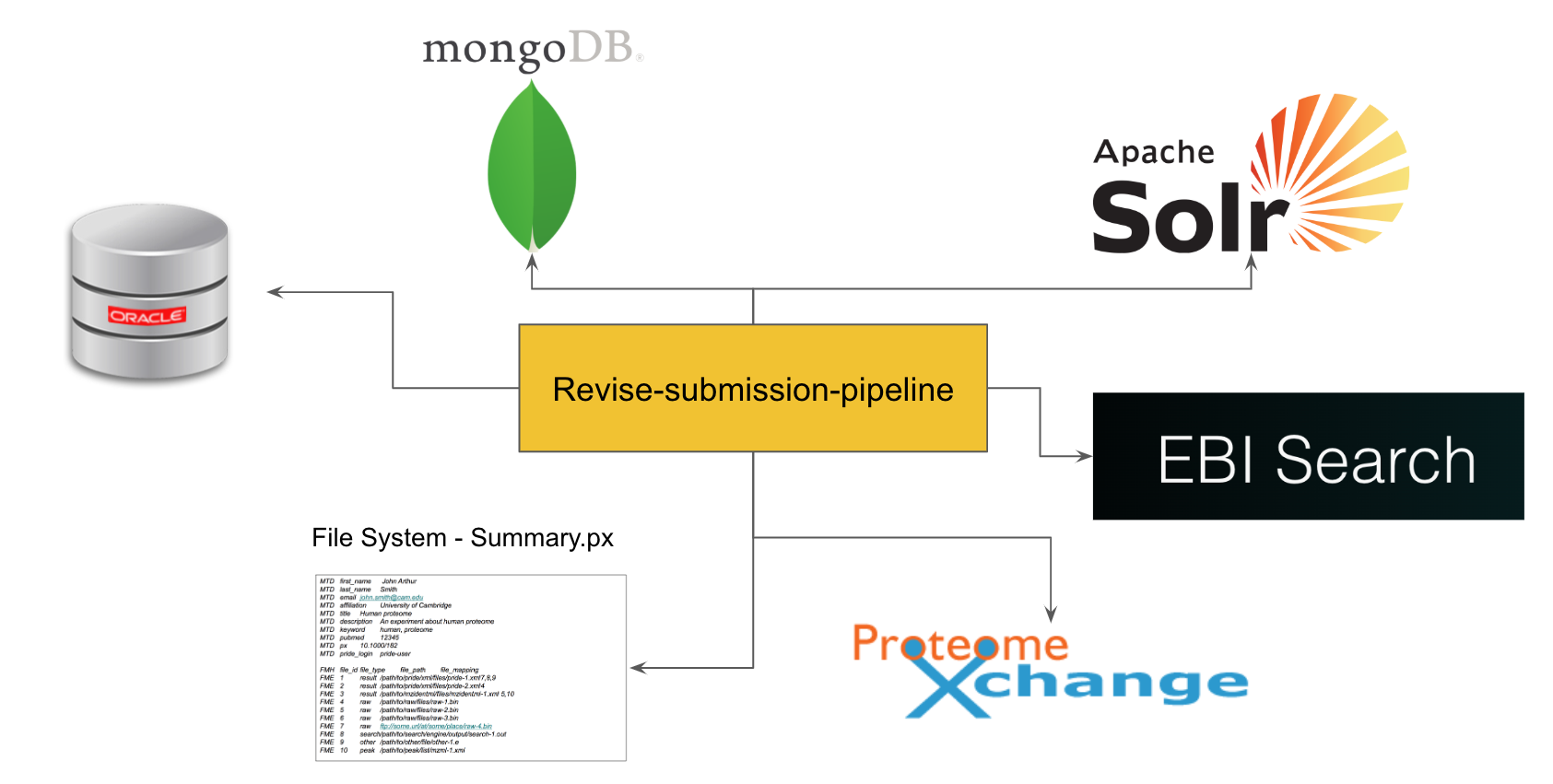
PRIDE Curation Tool - Reborn!
Reimplemented PRIDE Curation Tool to easily update metadata of projects in different services including MongoDB, Oracle, Solr, Redis, Flat file system etc.